Rows: 248,218
Columns: 58
$ id <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15,…
$ name <chr> "augmentin 625 duo tablet", "azithral 500 tablet",…
$ substitute0 <chr> "Penciclav 500 mg/125 mg Tablet", "Zithrocare 500m…
$ substitute1 <chr> "Moxikind-CV 625 Tablet", "Azax 500 Tablet", "Ambr…
$ substitute2 <chr> "Moxiforce-CV 625 Tablet", "Zady 500 Tablet", "Zer…
$ substitute3 <chr> "Fightox 625 Tablet", "Cazithro 500mg Tablet", "Ca…
$ substitute4 <chr> "Novamox CV 625mg Tablet", "Trulimax 500mg Tablet"…
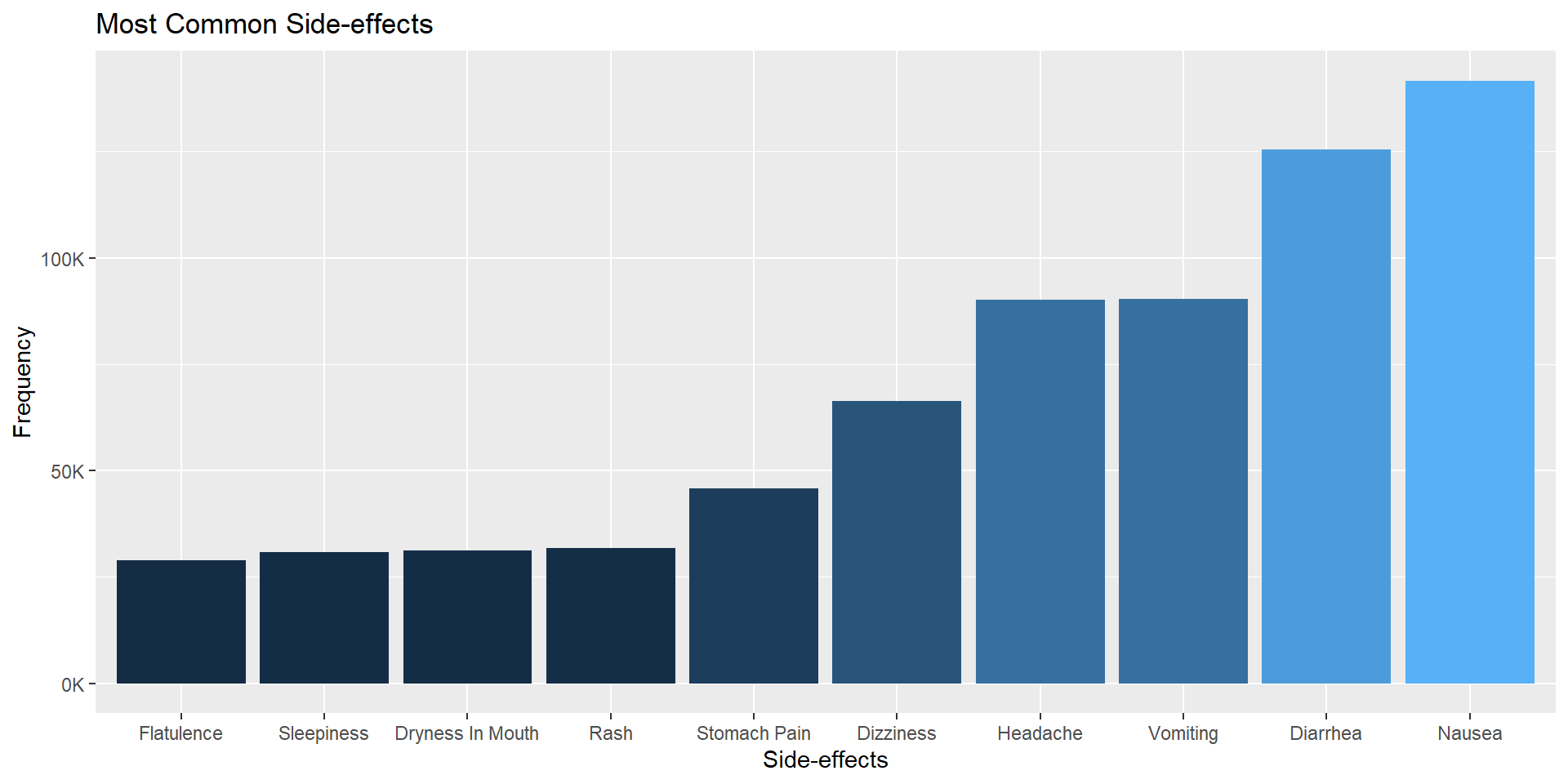
$ sideEffect0 <chr> "Vomiting", "Vomiting", "Nausea", "Headache", "Sle…
$ sideEffect1 <chr> "Nausea", "Nausea", "Vomiting", "Drowsiness", "Dry…
$ sideEffect2 <chr> "Diarrhea", "Abdominal pain", "Diarrhea", "Dizzine…
$ sideEffect3 <chr> NA, "Diarrhea", "Upset stomach", "Nausea", NA, "Sk…
$ sideEffect4 <chr> NA, NA, "Stomach pain", NA, NA, "Flu-like symptoms…
$ sideEffect5 <chr> NA, NA, "Allergic reaction", NA, NA, "Headache", N…
$ sideEffect6 <chr> NA, NA, "Dizziness", NA, NA, "Drowsiness", NA, NA,…
$ sideEffect7 <chr> NA, NA, "Headache", NA, NA, "Dizziness", NA, NA, N…
$ sideEffect8 <chr> NA, NA, "Rash", NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect9 <chr> NA, NA, "Hives", NA, NA, NA, NA, NA, NA, NA, NA, N…
$ sideEffect10 <chr> NA, NA, "Tremors", NA, NA, NA, NA, NA, NA, NA, NA,…
$ sideEffect11 <chr> NA, NA, "Palpitations", NA, NA, NA, NA, NA, NA, NA…
$ sideEffect12 <chr> NA, NA, "Muscle cramp", NA, NA, NA, NA, NA, NA, NA…
$ sideEffect13 <chr> NA, NA, "Increased heart rate", NA, NA, NA, NA, NA…
$ sideEffect14 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect15 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect16 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect17 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect18 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect19 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect20 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect21 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect22 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect23 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect24 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect25 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect26 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect27 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect28 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect29 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect30 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect31 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect32 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect33 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect34 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect35 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect36 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect37 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect38 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect39 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect40 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ sideEffect41 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ use0 <chr> "Treatment of Bacterial infections", "Treatment of…
$ use1 <chr> NA, NA, NA, "Treatment of Allergic conditions", NA…
$ use2 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ use3 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ use4 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
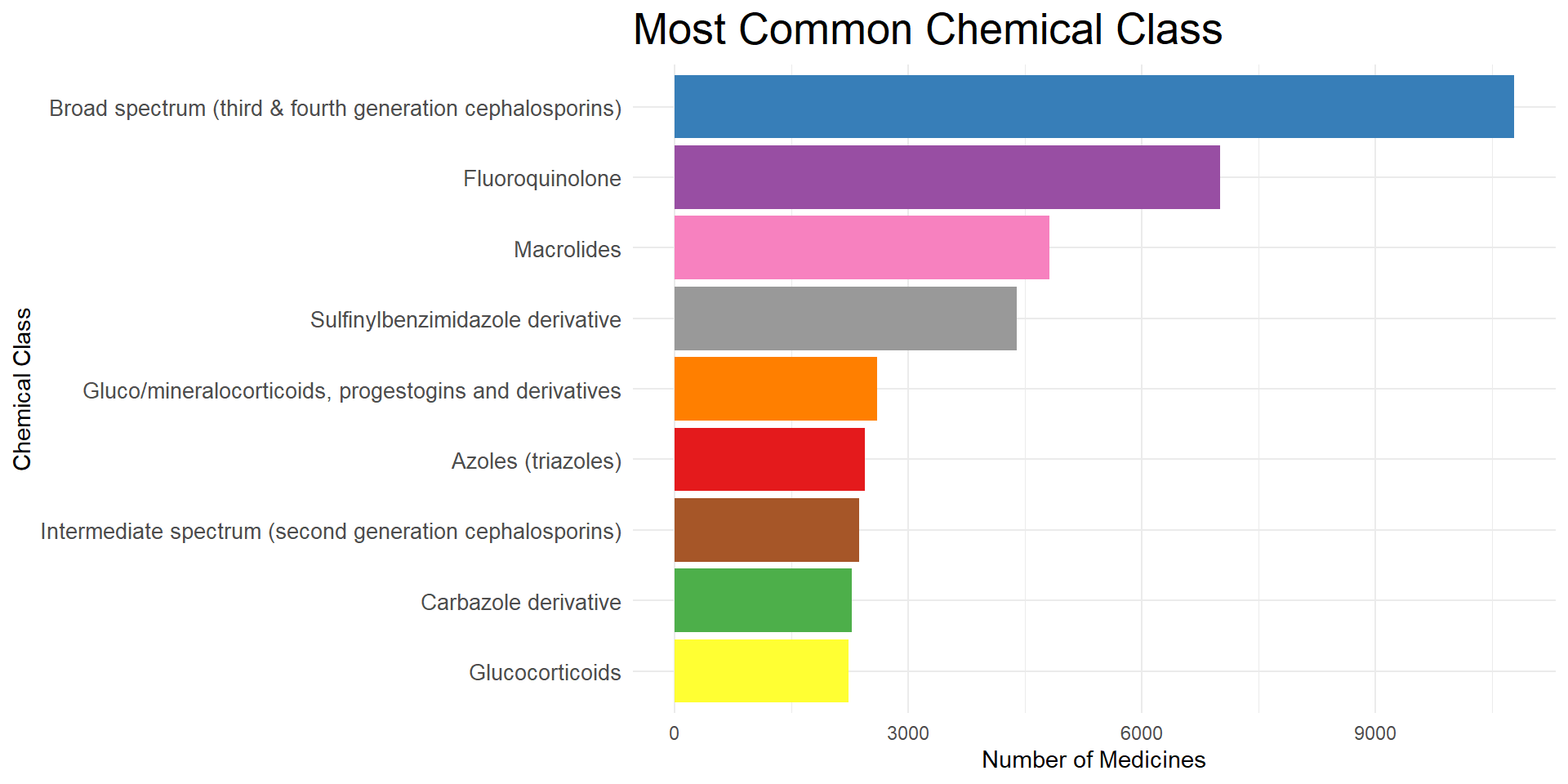
$ `Chemical Class` <chr> NA, "Macrolides", NA, "Diphenylmethane Derivative"…
$ `Habit Forming` <chr> "No", "No", "No", "No", "No", "No", "No", "No", "N…
$ `Therapeutic Class` <chr> "ANTI INFECTIVES", "ANTI INFECTIVES", "RESPIRATORY…
$ `Action Class` <chr> NA, "Macrolides", NA, "H1 Antihistaminics (second …