Predicting Diamonds Price
Introduction
Building a model to predict the price of the diamonds using tidymodels.
Diamonds data set is readily available to use through the ggplot2 library in the tidyverse and we will be using this data set predict the prices of the other diamonds.
In the data set various parameters of diamonds are given and each of these parameters may or may not effect the price of the diamonds.
library (tidyverse)library (plotly)library (DT)data ("diamonds" )
# A tibble: 53,940 × 10
carat cut color clarity depth table price x y z
<dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43
2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31
3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31
4 0.29 Premium I VS2 62.4 58 334 4.2 4.23 2.63
5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75
6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48
7 0.24 Very Good I VVS1 62.3 57 336 3.95 3.98 2.47
8 0.26 Very Good H SI1 61.9 55 337 4.07 4.11 2.53
9 0.22 Fair E VS2 65.1 61 337 3.87 3.78 2.49
10 0.23 Very Good H VS1 59.4 61 338 4 4.05 2.39
# ℹ 53,930 more rows
Data has over 50,000 observations which is good for modeling.
Exploring the data
The diamonds data set is available to explore in ggplot2 library as mentioned above.
Let’s check for NA’s before exploring the data
%>% map ( ~ sum (is.na (.))) %>% unlist ()
carat cut color clarity depth table price x y z
0 0 0 0 0 0 0 0 0 0
It’s really good that there are no NA’s but we have to be careful of the 0 in the numeric columns.
%>% select (carat, x, y, z) %>% arrange (x, y, z)
# A tibble: 53,940 × 4
carat x y z
<dbl> <dbl> <dbl> <dbl>
1 1 0 0 0
2 1.14 0 0 0
3 1.56 0 0 0
4 1.2 0 0 0
5 2.25 0 0 0
6 0.71 0 0 0
7 0.71 0 0 0
8 1.07 0 6.62 0
9 0.2 3.73 3.68 2.31
10 0.2 3.73 3.71 2.33
# ℹ 53,930 more rows
Diamonds cannot have a x(length), y(width), z(depth) of 0 and have weight. So let’s replace these values with NA or we can remove them out completely too.
%>% mutate (x = if_else (x == "0" , NA , x),y = if_else (y == "0" , NA , y),z = if_else (y == "0" , NA , z)) %>% datatable ()
Now lets visualize the distribution of the diamonds.
<- diamonds %>% ggplot (aes (carat)) + geom_freqpoly (binwidth = 0.05 )ggplotly (frequency_poly)
From the Figure 1 we can observe that - Most of the diamonds are between 0.2 to 1.5 carats. - There are peaks which means higher number of diamonds at whole and common fractions.
My general knowledge is that the weight i.e, carat of the diamond influences the price most. Let’s visualize that.
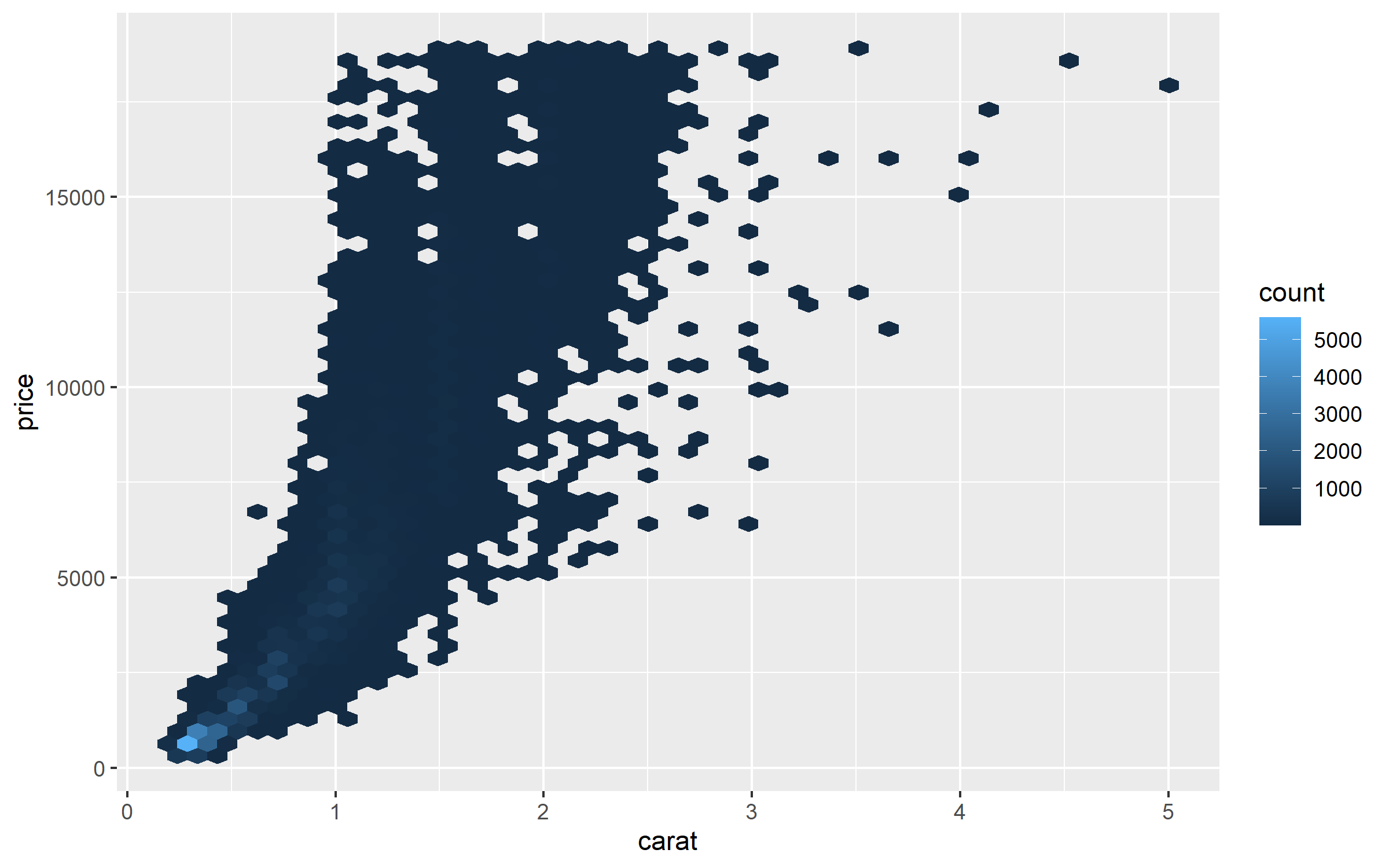
%>% ggplot (aes (carat, price)) + geom_hex (bins = 50 )
The price tends to follow exponential curve the log2() curve, we can confirm this by another graph.
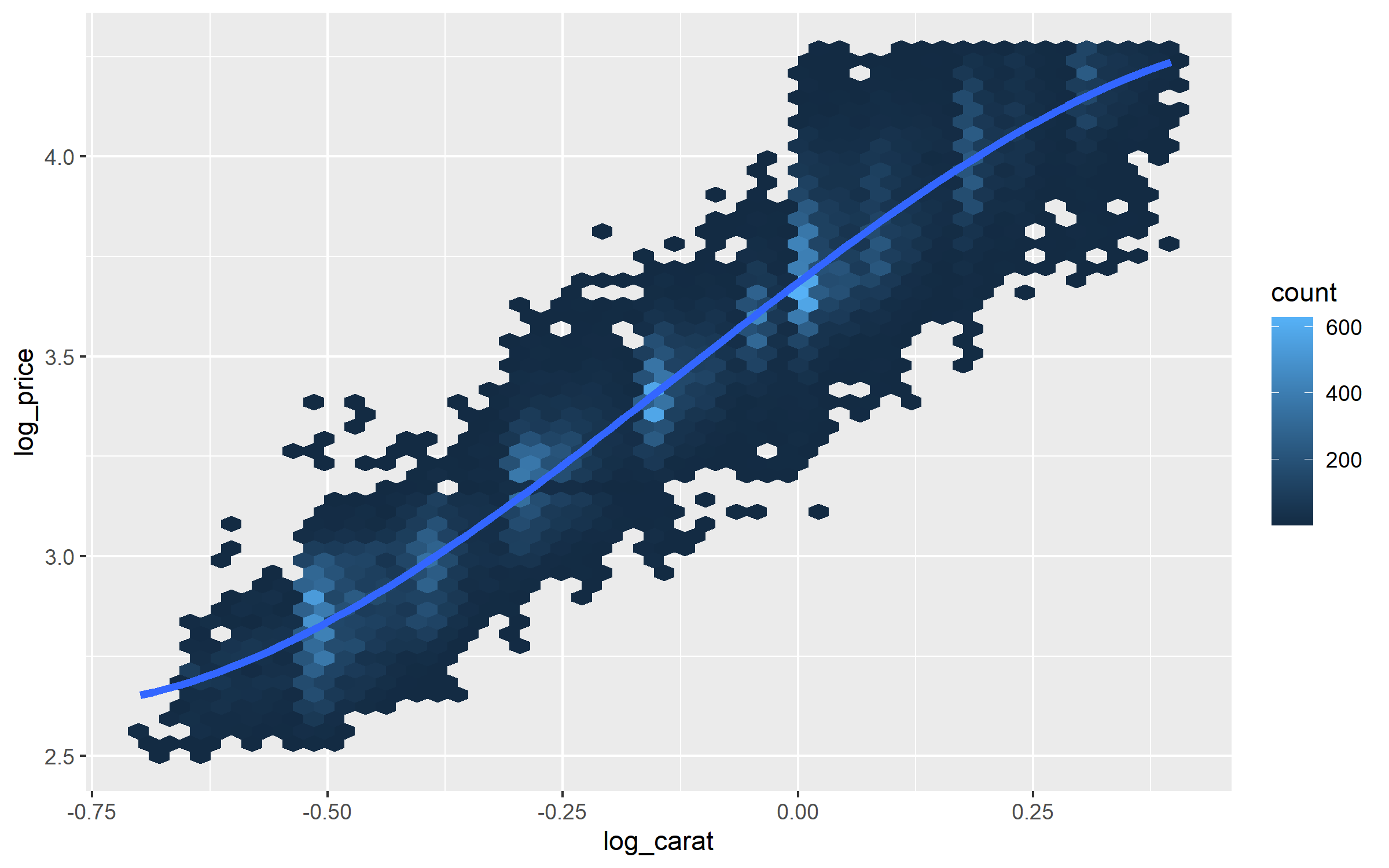
%>% filter (carat < 2.5 ) %>% mutate (log_price = log10 (price),log_carat = log10 (carat)) %>% ggplot (aes (log_carat, log_price)) + geom_hex (bins = 50 ) + geom_smooth (method = "lm" , formula = y ~ splines:: bs (x, 3 ),se = FALSE , linewidth = 1.5 )
The above Figure 2 shows that once we apply log2() to both price and carat the relationship mostly looks to be linear.
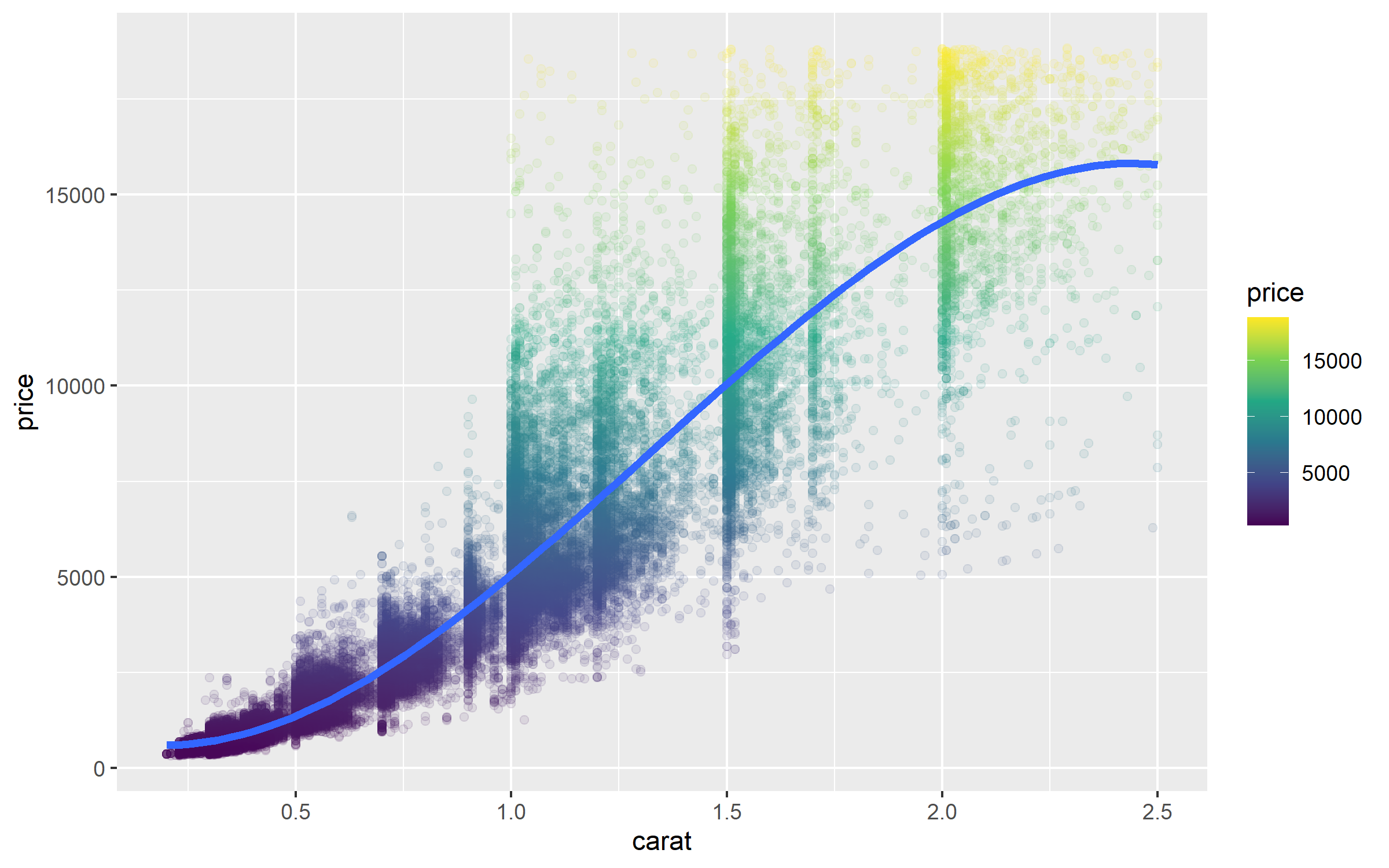
%>% filter (carat <= 2.5 ) %>% ggplot (aes (carat, price)) + geom_point (alpha = 0.1 , aes (color = price)) + geom_smooth (method = "lm" , formula = y ~ splines:: bs (x, 3 ),se = FALSE , linewidth = 1.5 ) + scale_color_viridis_c ()
We can see that price jumps when the weight is exactly or greater than to the whole and common fractions such as 0.5, 1.0, 1.5 and 2.
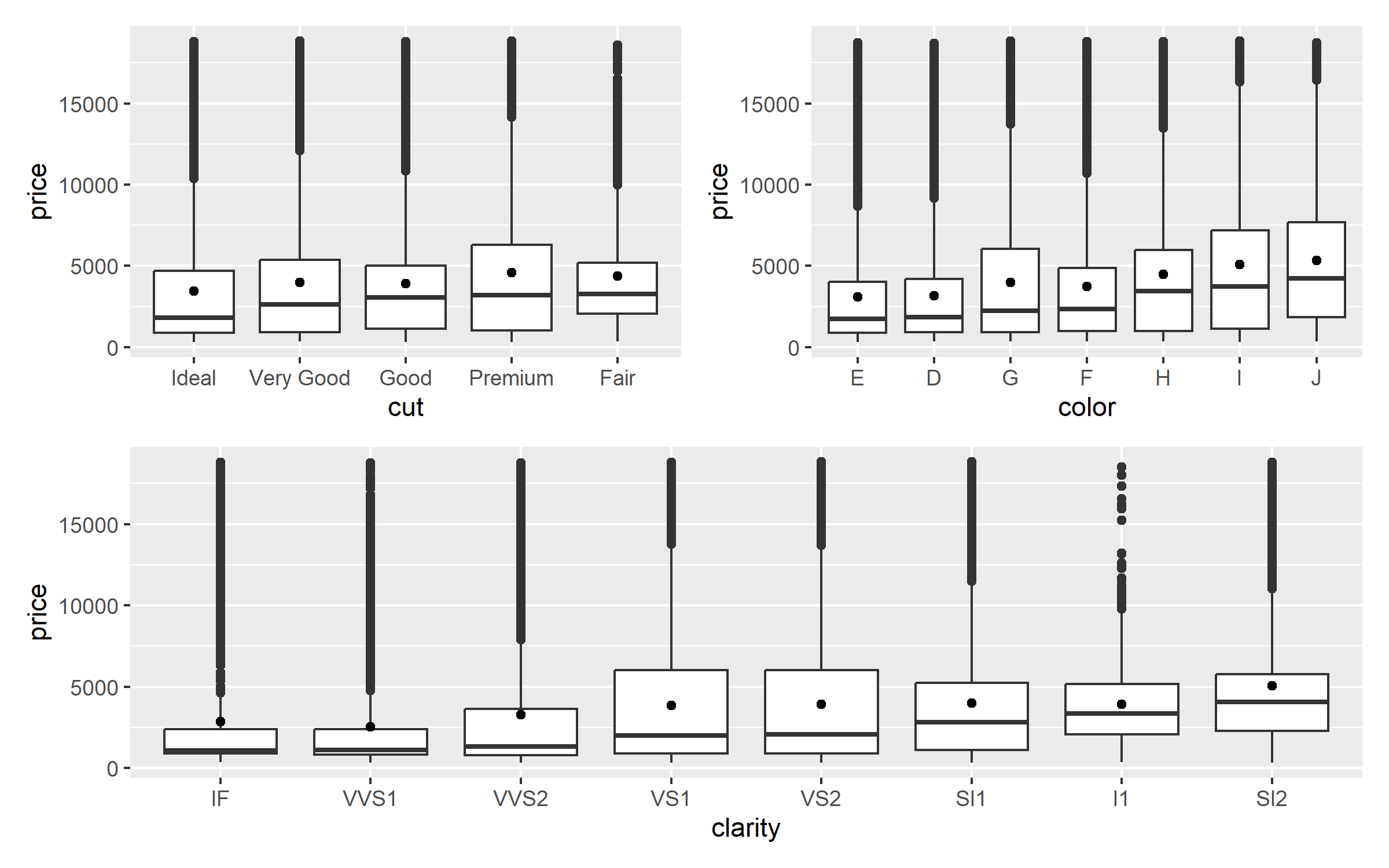
library (patchwork)<- function (param){ggplot (diamonds, aes (fct_reorder ({{param}}, price), price)) + geom_boxplot () + stat_summary (fun = mean, geom = "point" ) + labs (x = as_label (substitute (param)))plot_parameter (cut) + plot_parameter (color)) / plot_parameter (clarity))
Low quality diamonds with Fair cut and low quality color seems to have very high price. So now lets use tidymodels to model the data using rand_forest
Building a model
As every parameter in the data is important for the price prediction we are going to keep all the columns intact.
library (tidymodels)set.seed (2023 )<- diamonds %>% select (- depth) %>% mutate (price = log2 (price), carat = log2 (carat))<- initial_split (diamonds_2, strata = carat, prop = 0.8 )
<Training/Testing/Total>
<43150/10790/53940>
<- training (diamonds_split)<- testing (diamonds_split)
I am using strata with carat as most of the diamonds are not properly distributed yet all diamonds of different weight should be well represented.
<- vfold_cv (diamonds_train, strata = carat)
# 10-fold cross-validation using stratification
# A tibble: 10 × 2
splits id
<list> <chr>
1 <split [38833/4317]> Fold01
2 <split [38833/4317]> Fold02
3 <split [38834/4316]> Fold03
4 <split [38835/4315]> Fold04
5 <split [38835/4315]> Fold05
6 <split [38835/4315]> Fold06
7 <split [38836/4314]> Fold07
8 <split [38836/4314]> Fold08
9 <split [38836/4314]> Fold09
10 <split [38837/4313]> Fold10
I think rand_forest will work better on this data set but lets compare both Linear Regression models and Random Forest models.
<- linear_reg () %>% set_engine ("glm" )
Linear Regression Model Specification (regression)
Computational engine: glm
<- rand_forest (trees = 1000 ) %>% set_mode ("regression" ) %>% set_engine ("ranger" )
Random Forest Model Specification (regression)
Main Arguments:
trees = 1000
Computational engine: ranger
We still need to maniplulate some parts of the data like price and carat so that they are optimised which can be done using recipe library.
<- recipe (price ~ ., data = diamonds_train) %>% step_normalize (all_numeric_predictors ())<- base_recp %>% step_dummy (all_nominal_predictors ())<- ind_recp %>% step_bs (carat)
Next let’s start putting together a tidymodels workflow(), a helper object to help manage modeling pipelines with pieces that fit together like Lego blocks.
<- workflow_set (list (base_recp, ind_recp, spline_recp),list (lm_spec, rf_spec))
# A workflow set/tibble: 6 × 4
wflow_id info option result
<chr> <list> <list> <list>
1 recipe_1_linear_reg <tibble [1 × 4]> <opts[0]> <list [0]>
2 recipe_1_rand_forest <tibble [1 × 4]> <opts[0]> <list [0]>
3 recipe_2_linear_reg <tibble [1 × 4]> <opts[0]> <list [0]>
4 recipe_2_rand_forest <tibble [1 × 4]> <opts[0]> <list [0]>
5 recipe_3_linear_reg <tibble [1 × 4]> <opts[0]> <list [0]>
6 recipe_3_rand_forest <tibble [1 × 4]> <opts[0]> <list [0]>
Let’s fit the two models we prepared for the data. First code block contains linear regression model and the second contains the random_forest model.
:: registerDoParallel ()<- workflow_map ("fit_resamples" ,resamples = diamonds_folds
# A workflow set/tibble: 6 × 4
wflow_id info option result
<chr> <list> <list> <list>
1 recipe_1_linear_reg <tibble [1 × 4]> <opts[1]> <rsmp[+]>
2 recipe_1_rand_forest <tibble [1 × 4]> <opts[1]> <rsmp[+]>
3 recipe_2_linear_reg <tibble [1 × 4]> <opts[1]> <rsmp[+]>
4 recipe_2_rand_forest <tibble [1 × 4]> <opts[1]> <rsmp[+]>
5 recipe_3_linear_reg <tibble [1 × 4]> <opts[1]> <rsmp[+]>
6 recipe_3_rand_forest <tibble [1 × 4]> <opts[1]> <rsmp[+]>
Evaluating a model
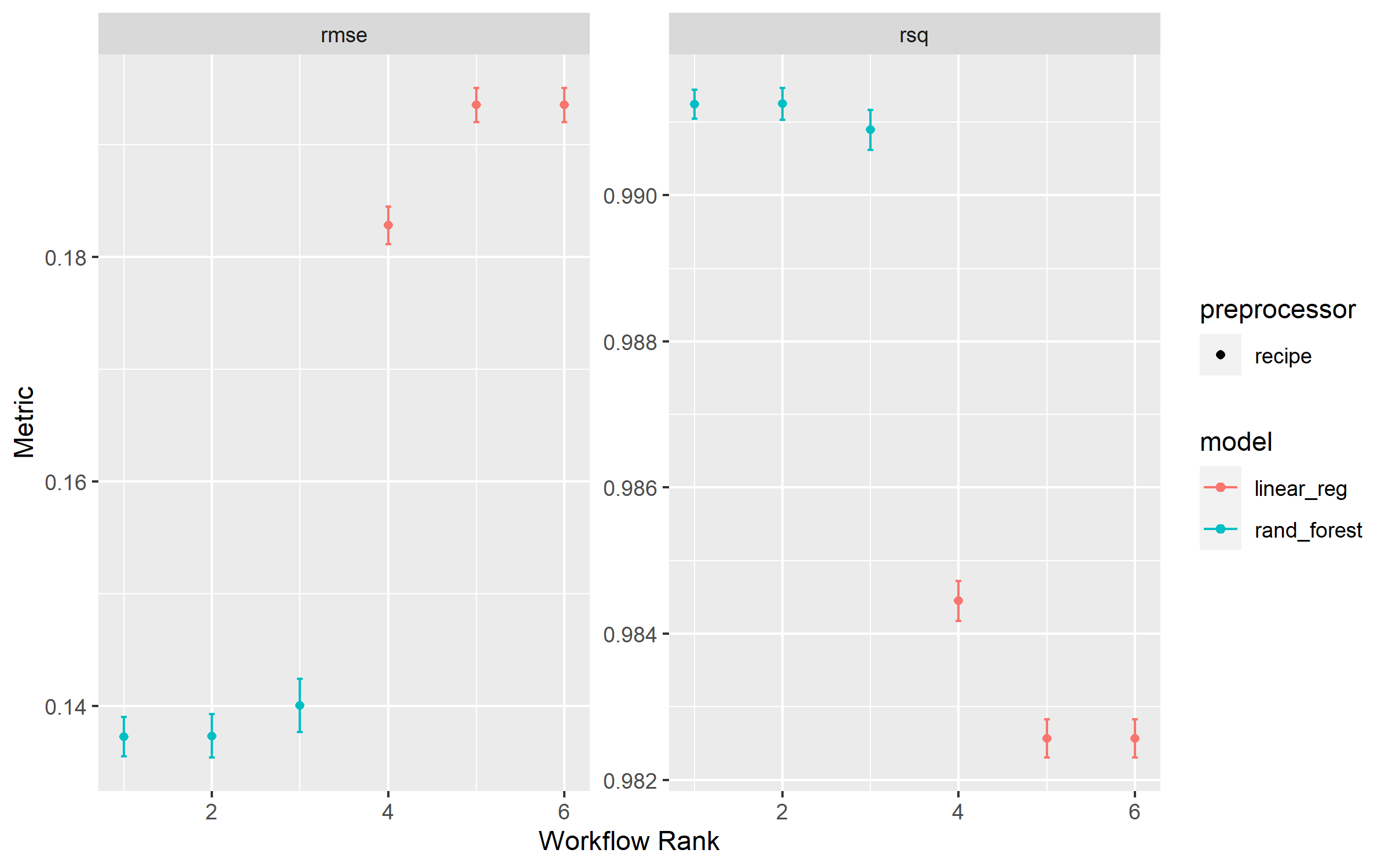
We can evaluate model by using autoplot and collect_metrics functions.
In the plot it seems that the difference between the rand_forest and linear_reg is very high but when we look at the metrics table we realise it’s not that much.
collect_metrics (diamonds_rs)
# A tibble: 12 × 9
wflow_id .config preproc model .metric .estimator mean n std_err
<chr> <chr> <chr> <chr> <chr> <chr> <dbl> <int> <dbl>
1 recipe_1_linear… Prepro… recipe line… rmse standard 0.194 10 9.25e-4
2 recipe_1_linear… Prepro… recipe line… rsq standard 0.983 10 1.58e-4
3 recipe_1_rand_f… Prepro… recipe rand… rmse standard 0.137 10 1.17e-3
4 recipe_1_rand_f… Prepro… recipe rand… rsq standard 0.991 10 1.33e-4
5 recipe_2_linear… Prepro… recipe line… rmse standard 0.194 10 9.25e-4
6 recipe_2_linear… Prepro… recipe line… rsq standard 0.983 10 1.58e-4
7 recipe_2_rand_f… Prepro… recipe rand… rmse standard 0.140 10 1.44e-3
8 recipe_2_rand_f… Prepro… recipe rand… rsq standard 0.991 10 1.65e-4
9 recipe_3_linear… Prepro… recipe line… rmse standard 0.183 10 1.01e-3
10 recipe_3_linear… Prepro… recipe line… rsq standard 0.984 10 1.67e-4
11 recipe_3_rand_f… Prepro… recipe rand… rmse standard 0.137 10 1.06e-3
12 recipe_3_rand_f… Prepro… recipe rand… rsq standard 0.991 10 1.19e-4
From the metrics table we can see that recipe_1_rand_forest seems to perform the best.
<- extract_workflow (diamonds_rs, "recipe_1_rand_forest" ) %>% fit (diamonds_train)<- pull_workflow_fit (final_fit)
parsnip model object
Ranger result
Call:
ranger::ranger(x = maybe_data_frame(x), y = y, num.trees = ~1000, num.threads = 1, verbose = FALSE, seed = sample.int(10^5, 1))
Type: Regression
Number of trees: 1000
Sample size: 43150
Number of independent variables: 8
Mtry: 2
Target node size: 5
Variable importance mode: none
Splitrule: variance
OOB prediction error (MSE): 0.01859615
R squared (OOB): 0.9913472
Let’s fit the test set to the model.
<- predict (object = final_fit,new_data = diamonds_test)
# A tibble: 10,790 × 1
.pred
<dbl>
1 8.85
2 8.64
3 8.66
4 8.72
5 8.70
6 8.62
7 8.87
8 8.73
9 8.83
10 8.63
# ℹ 10,780 more rows